Table of Contents
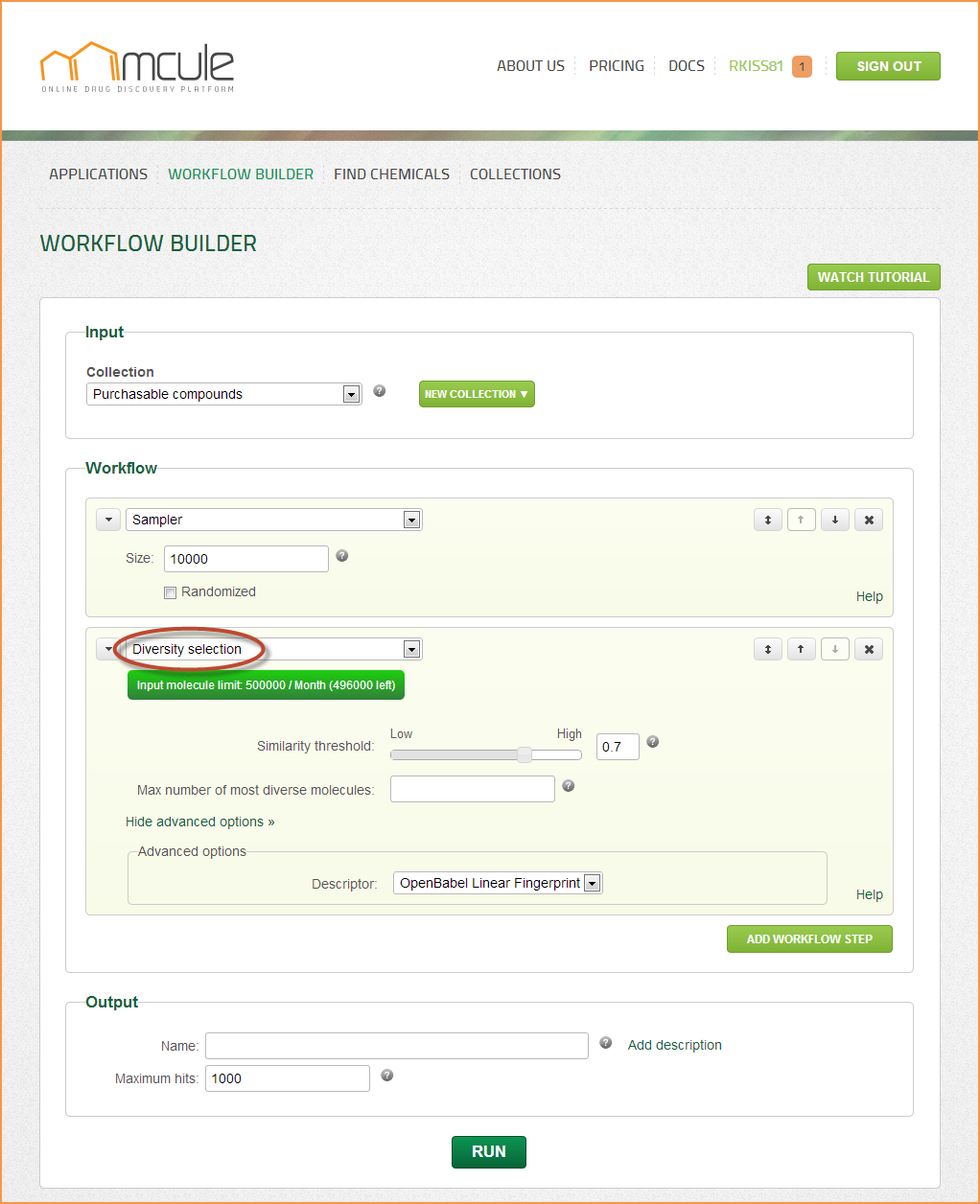
Diversity selection
This filter selects the most diverse (dissimilar) molecules from collections by eliminating the closest analogs. Diversity selection reduces the size of the input collection and maximizes the coverage of the chemical space at the same time.
When to use
If you have limited experimental or computational resources, diversity selection is an unbiased way to limit the number of compounds to handle. Collecting compounds from different regions of the chemical space is an efficient strategy to maximize the diversity of the identified active scaffolds.
Using this filter you can either reduce the size of large (virtual) screening libraries, or select a diverse, representative set of your virtual hits.
How to use
Exotic (non-druglike) molecules typically show large structural diversity. Consequently, they might be over-represented in the output of the Diversity selection filter. It is therefore recommended to eliminate such molecules prior to Diversity selection e.g. by the REOS filter.
Basic options
Similarity threshold
No molecule pairs in the output collection can be more similar to each other than this number (Tanimoto coefficient).
Max number of most diverse molecules
The diversity selection algorithm will output this number of molecules or less (if the “Similarity threshold” is reached first).
Advanced options
Descriptor
Mathematical representation of molecules used in the similarity/dissimilarity calculation. The The OpenBabel Linear Fingerprint (referred to FP2), and the Indigo Similarity Fingerprint (referred to sim) can be selected.
Tanimoto coefficient, the most widely used similarity metric, has been implemented and used as similarity measure by default.
Results
- diverse subset of the input collection satisfying the filter criteria
- maximum similarity (found between any molecule pairs if the rank list is cut exactly after this molecule) will be displayed as a single column in Table and List views
- output molecules will be ordered by maximum similarity (most diverse molecules are ranked highest)
Limits
Diversity selection available in the Free package is limited to 10,000 input molecules per month. To get access to unlimited Diversity selection, subscribe to our Library Design package.
Algorithm
Diversity selection utilizes an optimized implementation of the stepwise elimination algorithm (R. J. Taylor, J. Chem. Inf. Comput. Sci., 1995, 35, 59-67.), which can be described as follows:
- Calculate the similarity matrix of the molecules in the input collection
- Process the matrix elements as follows:
- Select the largest off-diagonal element in the similarity matrix
- Eliminates one molecule of the most similar molecule pair randomly
- Go to step 1. if off-diagonal elements remained
- Sort the list of eliminated molecules by similarity values associated to the elimination steps in increasing order
During this process, the size of the collection is reduced while the diversity of the collection is increased. Each elimination step filters out one molecule that has close analogues in the remaining set. As a result, the remaining molecules will have a decreased similarity (increased diversity).