Table of Contents
Mcule Advanced Curation (MAC)
The mcule database is curated by MAC (Mcule Advanced Curation) that involves a rigorous molecule registration system based on more than 80 structural checks, standardization, preparation and correction steps. MAC guarantees high quality search results and avoids common errors arising from mis-drawn and incorrect structures that can critically affect the quality of computational calculations and the efficiency of experimental results.
Key features of MAC: high level data curation, stereochemical standardization, robust novelty check and isomer detection, correct handling of salts & organometallics
Continue reading for more information about MAC, or check our presentations from the 244th National Meeting of American Chemical Society:
Evaluation of data quality in currently available compound libraries (slides)
Evaluation of data quality in currently available compound libraries (poster)
Quality is important
The design of screening libraries and the development of predictive drug discovery models all start with a high quality database. Chemical correctness is crucial because mis-drawn and imperfectly defined structures result in incorrect models, misleading predictions and inconsistent hits. Problematic structures should therefore be eliminated at the earliest possible stage from a drug discovery pipeline.
The mcule structure registration system is primarily designed to correctly handle chemical structures coming from different data sources, mainly from chemical suppliers, and load the structures into the mcule database. This is a non-trivial task which requires a careful structure check and preparation procedure. To reach a high curation level, the registration system should ensure database quality in terms of structure correctness, uniqueness and reliability as well as maintain a high level of data standardization.
All molecules with an MCULE ID have been processed by MAC. User uploaded molecules are not processed by MAC by default. We plan to enable this option in future.
Registration challenges
Standardization
Primary data sources of the mcule database are chemical supplier databases. Compounds from different supplier catalogs are often represented by different structure drawing standards. Some of these non-standard representations (e.g. salts, organometallic complexes and functional groups) can lead to difficulties e.g. during structure novelty check. Correct interpretation of suppliers’ stereochemical notations is also crucial. This is probably the most problematic area since the IUPAC stereo recommendations have been shown to be very difficult to implement in a cheminformatic system.
To interpret stereochemistry correctly, one should be aware of both the IUPAC convenction on stereo drawing and the stereo specification of SD file format used to store chemical structures by most chemical suppliers. Moreover, there are cases, where both of these conventions are violated, which the registration system also needs to be prepared to handle. Therefore, chemical data from different data sources should be very carefully analyzed, and stereo configurations need to be cleaned up to prevent the use of unreliable information and the misinterpretation of non-standard notations.
Need for data curation
Input structures might contain many kinds of problems. If these problems are not analyzed and errors are not corrected, misdrawn or insufficiently defined structures with e.g. incorrect valence states could enter the final database. It is important to note that not only should the obviously wrong structures be eliminated, but also the unreliable, or even potentially misdrawn ones. Considering the diversity of suppliers’ libraries and the number of potential problems that can arise, a carefully designed registration system is necessary to cope with as many error types as possible.
Checking structure novelty
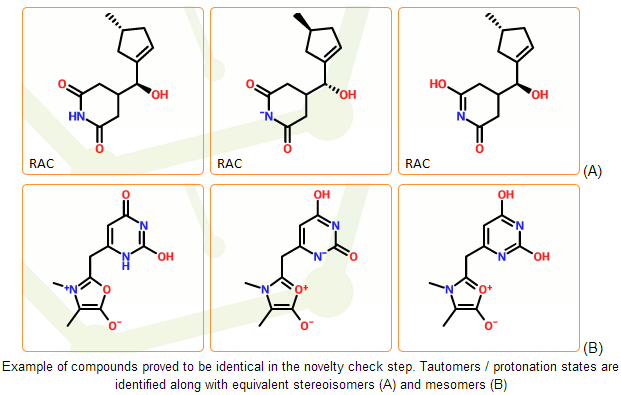
This is a very challenging task that cannot be solved perfectly in automated systems. Key problems are tautomerism and stereoisomerism.
Common forms of tautomerism can be detected with a rule-based system. But perceiving less common tautomer forms remains a problem even for experts. Stability of potential tautomeric forms cannot be well estimated, and the lack of appropriate computational methods can be only replaced with experience.
Correct handling of stereoisomerism is associated with correct tautomer detection. Without the correct identification of mobile hydrogens the symmetries of the structure can be underestimated. In addition, as the complexity of stereo representation increases the detection of identical isomers is getting more difficult. Here one is faced with a normalization problem.
Registration step types
The structure registration process involves many structure check & preparation steps, many novelty check algorithms, and a component separation algorithm in a fixed sequential order (not exactly in the order as listed in this documentation). The number of distinct registration steps in the system is more than 80.
Steps are primarily classified by their function. Structure check steps do not modify the structures, while preparation steps do. Preparations can be further grouped into (i) standardization, (ii) normalization and (iii) structure correction steps. Standardization steps modify the notations used to represent a given structure, while normalization steps keep the notations intact but do transformations between equivalent structures. These latter two preparation steps both uniformize structures and prepare them for the novelty check.
| Step | Function |
|---|---|
| Structure check | Prevent registration of incorrect, misdrawn or undesirable structures |
| Structure standardization | Change the notation system; structure representation is changed, structures are not |
| Structure normalization | Select a representative structure from equivalent structures; structure is changed, notation system is kept intact |
| Structure correction | Fixing some errors, remove problematic structural parts |
By design, the registration system not only filters out clearly wrong structures but also tries to detect potentially incorrect ones that cannot be handled automatically. The latter ones are not registered, and are awaiting for further manual correction and validation. Analyzing these registration cases can help in continuously improving our registration system. Adding new rules increases the level of automatization and decreases the need for manual curation.
Process outline
The whole registration process can be divided into seven different stages. It begins with the revision of stereo configurations, structure check/preparation steps (stage A, B) followed by component separation (stage C). Thereafter component uniqueness is checked and mcule IDs are assigned (stage D, E). This is performed with or without considering tautomerism and protonation, resulting the assignment of tautomer and protonation state independent compound identifiers (stage D) as well as tautomer and protonation state dependent structure identifiers (stage E). Finally, based on component identity, multicomponent entries are also registered at both the tautomer and protonation state independent (stage F) and dependent levels (stage G).
| Stage A | Enforcing standard stereo representation; non-standard stereo notations are changed, unreliable part of stereo configurations is removed (after consulting with chemical supplier) |
| Stage B | Product integrity and structure checks, functional group standardization, enforcing proper organometallic & salt representation, removing undesirable structures |
| Stage C | Common counterions are disconnected and disconnected components are separated |
| Stage D & E | Components are normalized, unique components are registered and new mcule IDs assigned at both compound (D) and structure (E) levels |
| Stage F & G | Full multicomponent structures are normalized, unique structures are registered with new mcule IDs assigned at both compound (F) and structure (G) levels |
As a result, input entries as well as their components are registered at two levels: tautomer and protonation state independent compound level with tautomer detection and tautomer and protonation state dependent structure level without tautomer detection.
Registration process
Stage A. Stereo clean up
Summary: enforcing standard stereo representation; non-standard stereo notations are corrected, unreliable part of the stereo configuration is removed
The registration system can be flexibly configured to interpret input stereo configurations according to the available information received from the chemical supplier. In the lack of information we keep only the reliable parts of the configuration and remove all the others.
During the registration of supplier catalogs or external libraries we always use the following procedure:
- we contact the compound supplier and inquire about the used stereochemical representation
- based on the answers we configure the registration system to use the proper stereo clean-up schema
- we try to store as many details about stereochemistry as possible, but keep reliable information only
- the applied stereo interpretation rules are always confirmed by the chemical supplier
- based on the clean-up schema, the registration system standardizes the stereo configuration, removes unreliable configurations and changes the stereo notations if needed
If no information is provided by the chemical supplier, we apply the following procedure:
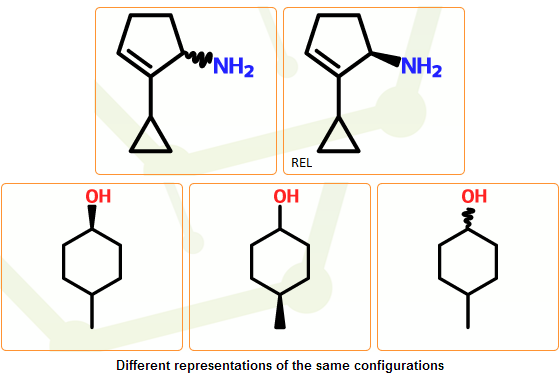
Cis-trans double bonds: all cis/trans configuration are marked as undefined: we assume that they can denote E (trans), Z (cis) configurations or the mixture of the two isomers.
Tetrahedral configuration: unmarked stereocenters or stereocenters marked with wavy bond(s) are treated as undefined, resulting the removal of the wavy bond. The stereo configuration type of the structure will be marked as “unknown”, indicating that it is not known whether the configuration is really absolute or not. It can be either relative or racemic, representing the apparent structure, its enantiomer or the racemic mixture of the two.
Stage B. General structure check & preparation
Summary: product integrity and structure checks, functional group standardization, enforce proper organometallic & salt representation
This registration stage aims the elimination/correction of problematic structures and the preparation of structures for subsequent steps, especially for component separation. Structures are considered to be problematic if they are chemically incorrect, uncertain/ambiguous, misdrawn or have missing components.
In this section, we list some of the most important check and preparation steps grouped by their purpose and complemented with some examples.
Constitution check
Formal charges and valence states are checked, cases where the placement of hydrogens is ambiguous are detected. In some cases our system requires explicit hydrogens, where the valence state cannot be determined automatically. This happens mainly with inorganic atoms.
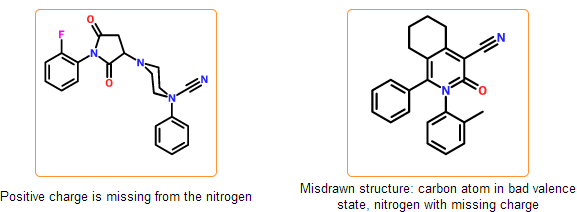
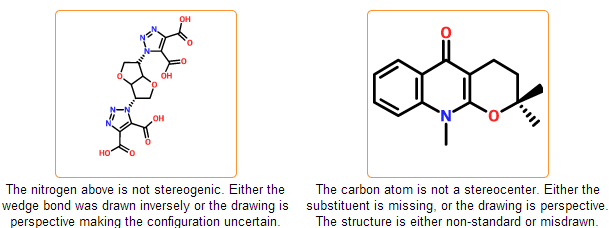
Configuration check
The stereo configuration in the input structures can be incorrect or ambiguous. In case of tetrahedral configuration wedge bonds denote the configuration around stereocenters. Wedge bonds can be problematic in the following cases:
- they are drawn to atoms that are not stereocenters
- they have wrong direction (wide end points to stereocenter)
- they are drawn between stereocenters indicating perspective drawing
- they have bad geometry
Similar problems can arise in case of cis/trans configurations that are also detected by the system. You can get further information about stereo drawing rules including the proper geometry of wedge bonds in the IUPAC documentation of Graphical Representation of Stereochemical Configuration.
Product integrity check
These registration steps detect misdrawn structures and check whether the input structures are completely specified. Certain registration steps are only performed when chemical supplier products are processed, as they should satisfy extra requirements.
One of the issues is that the input structure in the SDF can be incomplete (e.g. counterions are missing, stereo configuration is insufficiently specified, etc.). In these cases, additional information might be stored in the SDF fields (as data items) that affects the molecular structure. Such entries cannot be registered automatically, and are marked as problematic.
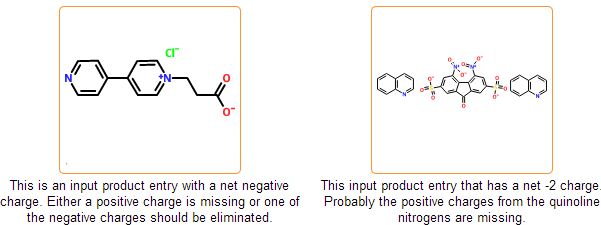
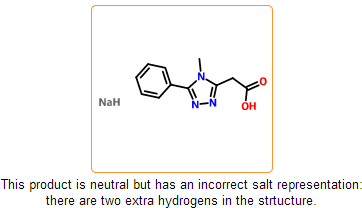
Even if the SDF is correct and all information is represented within the chemical structure, there is still a possibility that the structure is insufficiently specified or misdrawn. In certain cases we can detect such structures. For example, missing or extra hydrogens can be detected for special structural patterns. Moreover, purchasable product entries should have a net zero charge. Charged products usually indicates a missing or an extra counterion.
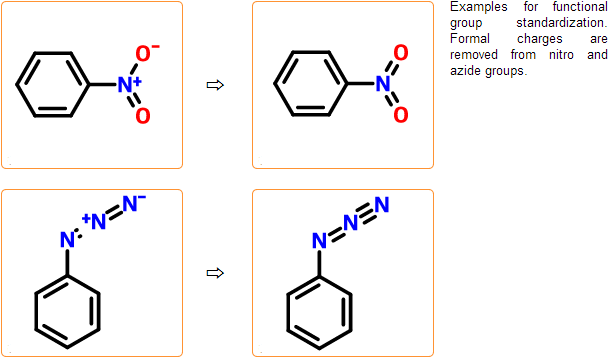
Functional group check & standardization
In these steps common functional groups such as nitro and azide groups are transformed to their neutral form. This standardization is necessary to get all relevant results from a substructure search. Besides standardization, several misdrawn forms of these functional groups are detected.
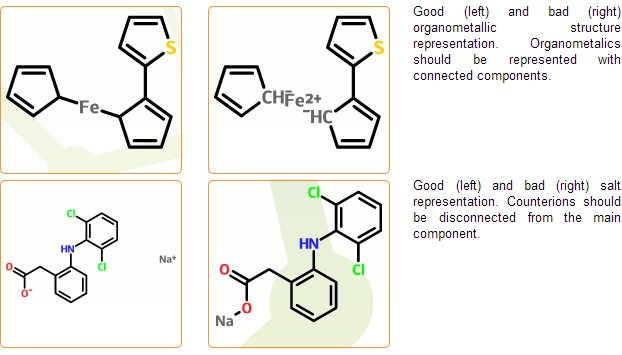
Enforce standard salt & organometallic compound representation
In the mcule database salts and organometallic complexes should be represented as disconnected and connected, respectively. Component separation is performed in the next registration stage where typical counterions are separated automatically. Cases, where salts cannot be distinguished from organometallic complexes cannot be processed automatically and are marked as problematic. Disconnected organometallic complexes where the reconnection of metals cannot be performed automatically are also marked as problematic.
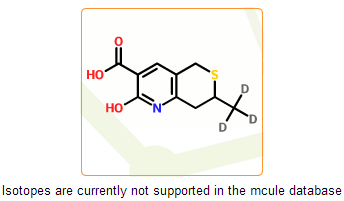
Elimination of undesirable structures
In the mcule database free radicals and isotopes are currently not supported. These structures have less relevance in drug discovery and are undesirable in virtual screening. Moreover, they are not supported well by some of cheminformatic tools that are implemented in the mcule system. In this step we detect such structures and prevent their registration.
SDF checks
We check SDF format and data consistency within the SDF entries. This step is necessary, since some data such as tetrahedral parities can be specified in SDF in multiple way. Furthermore only 2D structures are permitted, 0D or 3D ones are filtered out. They are stereochemically problematic and need extra treatment compared to 2D structures.
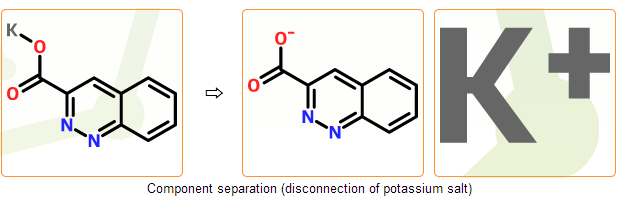
Stage C. Component separation
Summary: common counterions are disconnected and components are separated
In this stage we separate components of the incoming structure. In common salts counterions can be disconnected and separated from the main component automatically. Bonds to the main component are deleted and proper charges are placed on both components.
Stage D & E. Component registration
Summary: individual components’ structures are normalized, unique components are registered with new mcule IDs assigned at the tautomer and protonation state independent (D) and dependent (E) levels (steps in the D & E stages are very similar except for novelty check)
In stage D and E, components of the input structures are registered. Except for the novelty check, steps in the two branches are very similar, stereo normalization steps are followed by component novelty check. As a result, tautomer independent compound identifiers (short mcule IDs, stage D) and tautomer dependent structure identifiers (long mcule IDs, stage E) are assigned to the components.
Stereo configuration normalization
In the mcule system there are four stereo configuration types: absolute, relative, racemic and unknown (the “unknown” type is used to denote uncertain configurations, where compound provider could not confirm that the configuration type is really absolute). They are assigned in the stereo clean-up stage, and these initially assigned types are inherited by the separated components. In these steps these assigned stereo configuration types as well as the stereo configurations are further processed: for those components having no stereocenters, stereo configuration types are removed, while the stereo configuration of components with stereocenters are normalized together with their stereo configuration types.
Normalization is needed because certain configurations can be represented with multiple structures and/or stereo configuration types: replacing configurations around atoms and/or the configuration type can result in stereochemically equivalent structures. This can primarily happen when the configuration is only partially specified, containing atoms with both unknown/undefined and well-defined configurations. As a preparation step for the novelty check the same representative structures are selected from the set of structures with equivalent configurations.
Component novelty check
Main novelty check step is performed in stage D, focusing on the identification of different tautomer forms and protonation states of the same compound. In stage E different tautomers and protonation states are treated and registered as different structures.
In the mcule registration system the novelty check of the individual components is based on non-standard IUPAC InChI identifiers. The InChI software performs a lot of normalization steps and can detect common forms of tautomerism. It can also perceive protonation states of the same compounds in most cases. InChI strings therefore serve as a good starting point of novelty check. Different structures with identical InChIs can be considered as different representations of the same compound.
In stage D we use a novelty check algorithm that is based on the InChI strings but can detect an even broader set of potential tautomers than a simple InChI comparison. The system is capable to fully prevent the registration of duplicates as long as they are prototopic tautomers.
Stage E & F. Multicomponent structure registration
Summary: additional checks are performed, component types are assigned, and unique structures are registered with new mcule IDs assigned at the tautomer and protonation state independent (F) and dependent (G) levels (steps in the F & G stages are very similar)
Novelty check
Novelty check of multicomponent entries is based on component identity and component multiplicity. Two structures are treated as identical when they contain identical components with the same multiplicities.
This novelty check method needs some checks and preparations. The number of identical components (multiplicity) should be reduced to the lowest possible value, when they don’t store additional information. The presence of multiple components can indicate relative / racemic stereochemistry according to the IUPAC recommendations. These cases should be identified.
It is also important to identify contaminants. They should be eliminated and stored as a property of the product. Products with different contaminants are still related to the same compound.
Component type assignment
After novelty check components are analyzed. The system can identify counterions and potential solvents. In the latter case registration is problematic. For example, water can be a crystal water or can denote the solvent. Depending on its role it should be removed from or retained in the structure. Also, the deprotonated form of water (hydroxide) can serve as a counterion.
In most cases the system is also capable of identifying the main components, which can serve as the input set for virtual screens.
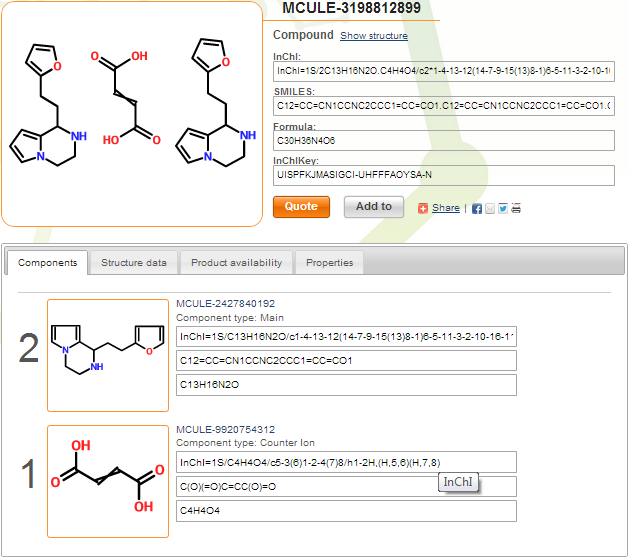
You can see below the index page of compound MCULE-3198812899. This is a maleic and/or fumaric acid salt (uncertainty is marked by crossed double bond). Counter ions are marked, and component multiplicities are assigned correctly by the system.