Table of Contents
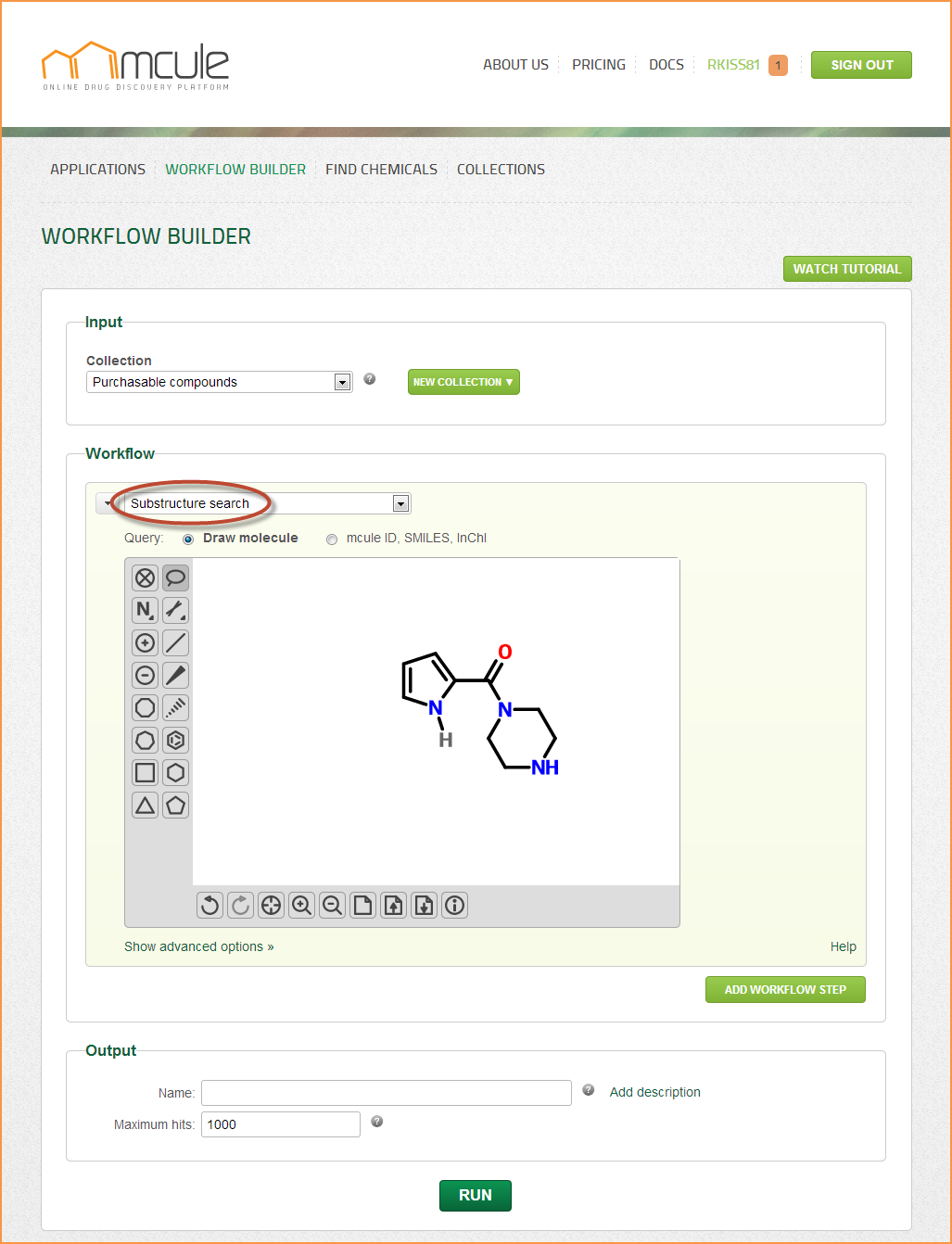
Substructure search
When to use
Substructure searching is frequently applied for hit exploration. Unlike similarity searching, it outputs only those molecules containing the whole query molecule. It can therefore retrieve close analogs of a particular scaffold (e.g. HTS hit or reference ligand), which can be used to build an initial SAR (structure-activity relationship).
How to use
If your first query does not yield enough hits, it is recommended to fine-tune your substructure search and use a more general query. If you get too many hits, you can block certain substitution positions by adding explicit hydrogens (see example in Figure). Multicomponent queries are currently not supported.
Option
It is possible to set whether the substructure search should be performed on the specific tautomer (and protonation state) specified in the input, or the search is tautomer (and protonation state) independent (“Tautomer independent” box).
Queries can be drawn by the molecule sketcher, which can be hidden and reopened by clicking on the “Hide/Show sketcher” link. Queries can be also defined by entering mcule IDs, InChI or SMILES strings into the input field. 2D representations can be generated by clicking on the “Generate 2D” button. Note: If the “Generate 2D” button was clicked, the search will be performed on the generated 2D representation (2D SDF exported from the sketcher). If, however, the “Generate 2D” button was not clicked, the search will be directly performed on the mcule ID, InChI or SMILES strings coming from the input field. The results might be slightly different in these two cases.
Results
Molecules satisfying search criteria