Table of Contents
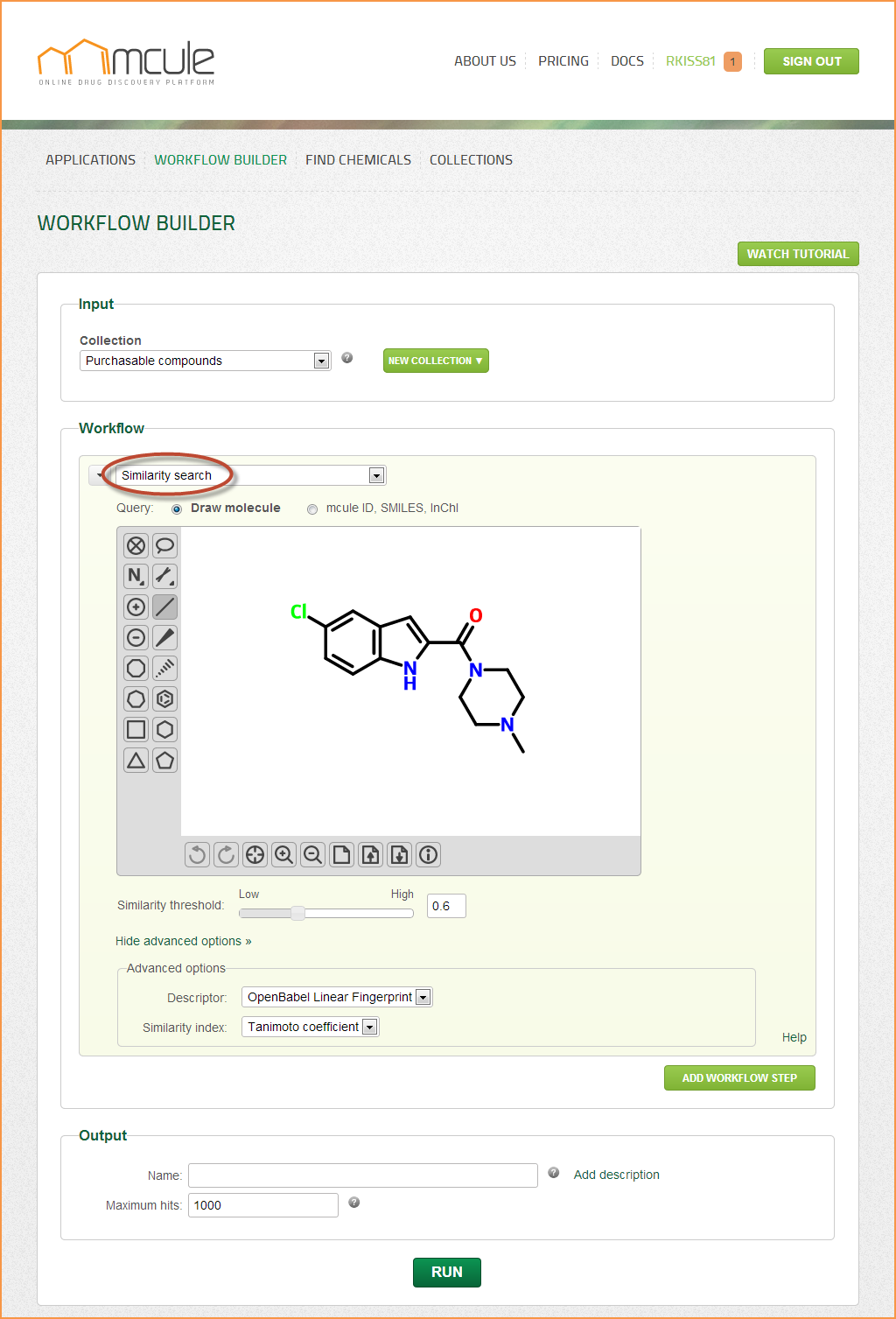
Similarity search
Similarity search is for finding structurally similar molecules to a query molecule.
Similarity between molecules can be defined in various ways. One typical approach, routinely applied in chemoinformatics is to represent the molecules with molecular fingerprints (descriptor) and calculate similarity by comparing the fingerprints rather than the actual molecular graphs. This allows very fast calculation of a simple similarity measure between molecules. Molecular fingerprints most frequently used for similarity searching store information about fragments and substructures present in the molecule. If the fragment is present, the corresponding bit in the fingerprint will be 1. If missing, the bit will be 0. These molecular fingerprints are essentially lists of 1s and 0s reducing the problem of molecular similarity calculation to comparison of bitstrings. The way we calculate a similarity value from the bitstrings is determined by the similarity metric.
When to use
Similarity search can be particularly useful for finding close analogs for a reference ligand or an initial hit identified by biological screening (e.g. HTS). Close analogs of actives are expected to show some activity (similarity paradigm) and they can be used to build an initial SAR (structure-activity relationship) around a scaffold.
How to use
Options
Descriptor
Mathematical representation of molecules used in the similarity/dissimilarity calculation. The OpenBabel Linear Fingerprint (referred to FP2), and the Indigo Similarity Fingerprint (referred to sim) can be selected.
Similarity metric
Tanimoto coefficient, the most widely used similarity metric, has been implemented and used by default.
Similarity threshold
The similarity threshold is the minimum similarity value between the query and target molecules (set to 0.6 by default). A similarity value of 1 means that the two molecules cannot be distinguished by the selected descriptor and similarity metric. If the similarity search does not yield any hits with the default threshold, it makes sense to set a lower threshold and try again.
Queries can be drawn by the molecule sketcher, which can be hidden and reopened by clicking on the “Hide/Show sketcher” link. Queries can be also defined by entering mcule IDs, InChI or SMILES strings into the input field. 2D representations can be generated by clicking on the “Generate 2D” button. Note: If the “Generate 2D” button was clicked, the search will be performed on the generated 2D representation (2D SDF exported from the sketcher). If, however, the “Generate 2D” button was not clicked, the search will be directly performed on the mcule ID, InChI or SMILES strings coming from the input field. The results might be slightly different in these two cases.
Results
- Molecules satisfying search criteria
- Tanimoto coefficient showing the similarity between the query and the target molecules will be displayed as a single column in Table and List views
- Hits will be ordered by similarity score